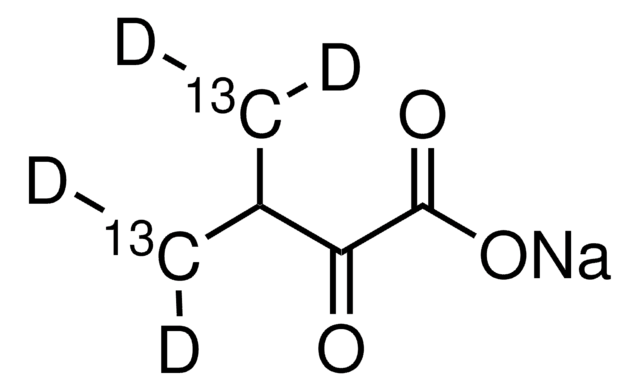
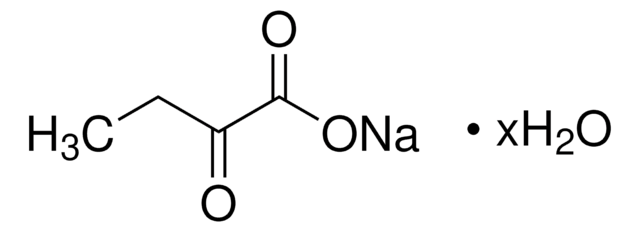
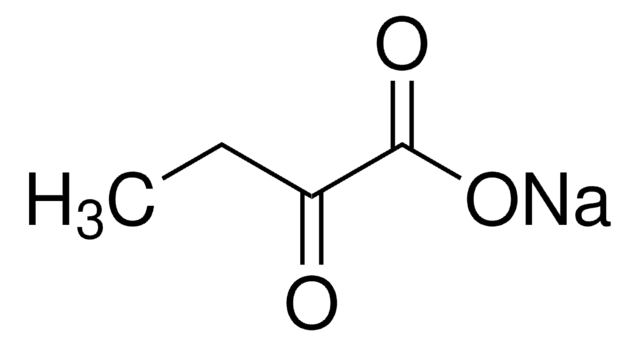
607541
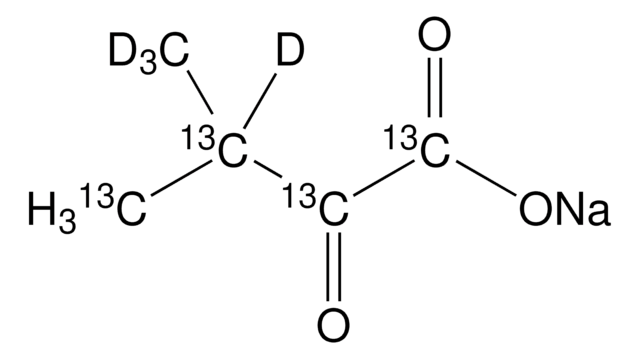
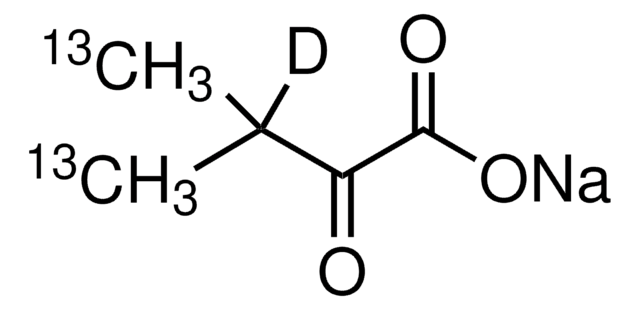
2-Ketobutyric acid-13C4,3,3-d2 sodium salt hydrate
99 atom % 13C, 98 atom % D, 98% (CP)
Synonym(s):
α-Keto-butyric acid-4-13C4-3,3-d2 sodium, α-Ketobutyric acid-4-13C4-3,3-d2 sodium salt, 2-Oxobutanoic acid-13C4,3,3-d2 sodium salt, Sodium α-ketobutyrate-13C4,3,3-d2
About This Item
Recommended Products
isotopic purity
99 atom % 13C
98 atom % D
Quality Level
assay
98% (CP)
form
solid
technique(s)
bio NMR: suitable
mp
210 °C (dec.) (lit.)
mass shift
M+6
SMILES string
O.[Na+].[2H][13C]([2H])([13CH3])[13C](=O)[13C]([O-])=O
InChI
1S/C4H6O3.Na.H2O/c1-2-3(5)4(6)7;;/h2H2,1H3,(H,6,7);;1H2/q;+1;/p-1/i1+1,2+1D2,3+1,4+1;;
InChI key
PVLJVKQSCAHPPR-WVMSAXHDSA-M
Related Categories
Packaging
wgk_germany
WGK 3
flash_point_f
Not applicable
flash_point_c
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Articles
Presenting an article about the selective protonation of methyl groups in highly deuterated proteins. In which the structural NMR studies of small proteins, a maximum number of proton chemical shifts are usually assigned and NOEs connecting large numbers of sites are subsequently quantified in terms of distance restraints that are then used to obtain an ensemble of structures.
We presents an informational article concerning biomolecular NMR and the use of Isotope Labeling Methods for Protein Dynamics Studies.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service