Microbiome Multi-omic Services
From sample collection to sequencing and metabolite identification, the Microbiome community has repeatedly shared the need for reproducible and translational worthy data to treat human and environmental diseases. Our Microbiome services are offering a multi-omic approach to customers globally who would like to outsource sequencing and metabolite identification of microbiome samples and receive a comprehensive and intuitive bioinformatic data report accessible on the cloud.
We are adding Microbiome multi-omic services to our Sigma-Aldrich® portfolio of products, to accommodate customer needs with varying biological and environmental samples and provide robust and comprehensive detection and analytical services from sample preparation to statistical reporting. We cater to various applications from 16S Amplicon sequencing for bacteria species classification to Shotgun Metagenomic sequencing to determine strain or sub-species level specificity and functional analyses. Our metabolic profiling services includes targeted identification of metabolite panels and untargeted screening.
With large and complex volumes of data, it is imperative to have simplicity and ease of use in a bioinformatics and statistic tool program, and hence the cloud-based Microbiome platform will permit novice to experienced users to analyze and generate publication ready figures for their research projects. The bioinformatics databases will be routinely updated as new species are identified and the cloud application will be web accessible, therefore voiding any requirements for an annual license or a software download. As part of the service offering, the project data generated will be uploaded to the cloud and a downloadable HTML report will be generated. This report is dynamic and interactive and can be emailed to collaborators as well as be cited in reference publications.
Our experts can aid in your clinical study design upstream for microbiome biologic sample collection preparation, statistical analysis and reporting downstream respectively over the course of your service project.
Request Information
The Microbiome platform encapsulates a multi-omics approach
- Metagenomics Services
- 16S Amplicon Sequencing
- Shotgun Metagenomic Sequencing
- SMURF sequencing
- Cloud based Metagenomic Platform for bioinformatics
- Service request and tracking of sample shipment and project status
- Standardized and up to date sequencing databases
- Intuitive bioinformatics software for 16S & Shotgun Metagenomic sequencing data files
- Metabolomics Services
- Targeted metabolite panel detection
- Untargeted metabolite screening
A Gut Instinct On Bioinformatics
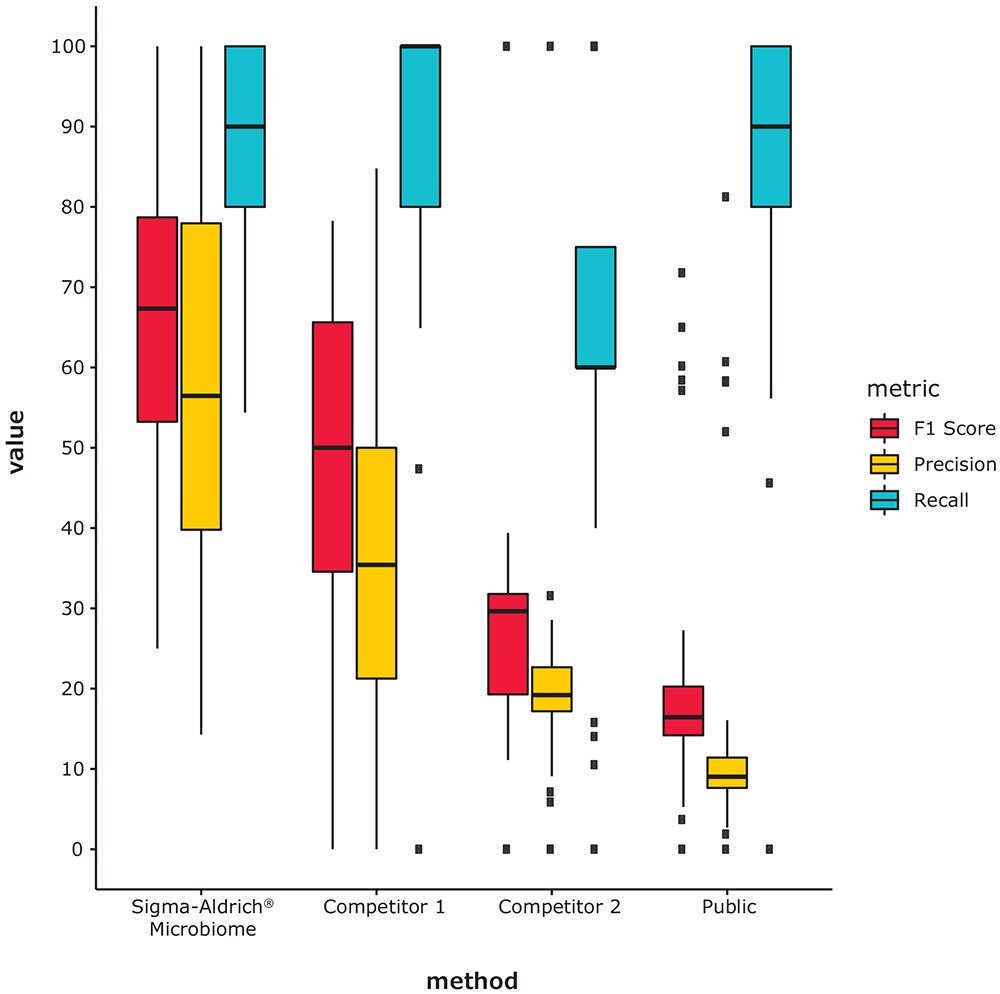
To compare our in-house Microbiome platform to competitors, 16S-seq standard mock community mixtures were used, including 23 samples sequenced by our Microbiome services laboratory and 9 published samples with publicly available Illumina MiSeq sequencing data1,2,3. We ran each sample through our Microbiome platform, and compared it to competing commercial cloud platforms, and an academic algorithm run with the our internally developed and curated 16S (V3-V4) database. The species-level results were compared using the F1 score, precision, and recall. Precision is inversely related to the number of false positive bacteria and recall is inversely related to the number of undetected true positive bacteria. The F1 score is a combination of precision and recall which gives an overall measure of how well the calculated and known taxonomic profiles match. The results show that our Microbiome platform provides robust performance in species-level profiling of complex microbial communities.
Microbiome Services Tracker
Purple: Customer; Cyan: Microbiome Service Lab
We wanted to provide you a simple and easy way to track the status of your service project, from the shipment of samples to our service lab, to the final data report upload and consultation with our experts. By setting up an account on our Microbiome Platform, you will be able to return to your project and tweak parameters that may have changed in the literature or QC requirements and re-run the classifications and comparative analyses.
References
To continue reading please sign in or create an account.
Don't Have An Account?